Product Description
The Gene-Foci® Site-directed mutagenesis kit is designed to introduce point mutations, replace, delete or add nucleotide(s), thus changing the amino acid sequence encoded by this fragment of DNA. The basic procedure utilizes PCR mediated by high-fidelity DNA polymerase, and a pair of primers that contains the mutation(s) wanted. After PCR amplification, the methylated template DNA is digested with the restriction enzyme DpnI, and the PCR product containing the mutation is transformed into DH5a competent cells, and positive clones can be selected by sequencing or other methods.
Features:
- Ultra high-fidelity DNA polymerase ensures the precise integration of point mutations and minimizes the chance of generating unwanted mutations.
- Capable of introducing mutations into long plasmid templates up to 16kb.
- Enhanced processivity of DNA polymerase ensures the efficiency of the PCR reaction.
- Super high-efficiency competent cells ensure the transformation of target plasmids.
Storage:
The DH5a ultracompetent cells should be stored at -80°C, all other components are stored at -20°C.
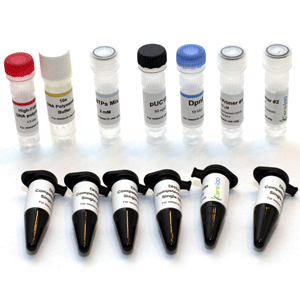
I. Kit components
|
Components |
Volume |
|
|
20 reactions #GF1011 |
6 reactions #GF1010 |
|
|
High-Fidelity DNA polymerase (2.5 U/μl) |
50 U |
20 U |
|
10x reaction buffer (with enhancers) |
300 µl |
200 µl |
|
dNTPs mix (2 mM each) |
40 µl |
20 µl |
|
DpnI (10 U/μl) |
20 µl |
8 µl |
|
pUC19 control plasmid (50 ng/µl) |
10 µl |
5 µl |
|
Control Primer #1(10 µM) |
20 µl |
10 µl |
|
Control Primer #2(10 µM) 5’-TCACACAGGAAACAGCTCTGACCGTGATTACGC-3’ |
20 µl |
10 µl |
|
DH5a ultracompetent cells (single shot) |
20 tubes |
8 tubes |
II. Primer Design hints
- Both the forward and the reverse complimentary primers should contain the desired mutation, and should anneal to the same locus on the original plasmid. You can design the forward primer, and then generate the reverse-complimentary sequence of it as the second primer.
- The length of the DNA oligos for use in the site-directed mutagenesis protocol should be between 32-40bp.
- The mutation site should be located near the center of the oligos to ensure efficient annealing to the template.
- HPLC or PAGE purification of primers is recommended.
III. Prepare mutagenesis PCR reaction
Prepare the reaction in 0.2 ml thin wall PCR tubes as indicated below:
|
Template plasmid* (100 ng/μl) |
0.5 μl |
|
10x reaction buffer (with enhancers) |
2 µl |
|
dNTPs mix (2 mM each) |
2 µl |
|
Primer #1 (10 μM) |
2 µl |
|
Primer #2 (10 μM) |
2 µl |
|
High-Fidelity DNA polymerase (2.5 U/μl) |
0.5 μl |
|
ddH2O |
Up to 20 μl |
* Plasmids purified from most of the commonly use E.coli strains (such as DH5a, BL21 etc.) are methylated, and are suitable for being used as template in site-directed mutagenesis. However, plasmid purified from dam– E.coli strains, such as JM110, should not be used as template without an appropriate in vitro methylation procedure.
Mix components in the tube by gently pipetting up and down several times. Briefly spin down, then proceed to step IV.
IV. Thermocycling program
Set up PCR program on a thermocycler as below:
|
Cycles |
Temperature |
Time |
|
| (1) |
1 |
95°C |
3 minutes |
| (2) |
16~18 |
95°C |
30 seconds |
|
55°C(a) |
30 seconds |
||
|
68°C |
1~2 min/kb(b) |
||
| (3) |
1 |
72°C |
10 minutes |
| (4) |
1 |
4°C |
Hold |
(a): The annealing temperature may need optimization for different DNA oligo primers.
(b): 2 minutes / kb is recommended for GC-rich or other difficult to amplify DNA sequences.
V. DpnI digestion of template DNA
After the PCR reaction is done, directly add 1 μl of 10 U/μl DpnI into the reaction tube, gently mix well, then incubate at 37°C for 1 hour. Purification of PCR product is not necessary.
(Optional) determine the DpnI digestion efficiency by digesting template plasmid-only control:
In a 0.2 ml thin-wall PCR tube, add:
Template plasmid* (100 ng/μl) 0.5 μl
10x reaction buffer (with enhancers) 2 μl
DpnI (10 U/μl) 1 μl
H2O 17.5 μl
Total volume: 21 μl
Incubate at 37°C for 1 hour. If digestion is complete, no colonies should result from transformation into Dh5a.
VI. Transformation of Dh5a competent cells
Before start:
- Set the temperature of a water bath to 42°C.
- Pre-warm LB media to 37°C
Transformation:
- Gently remove the single shot Dh5a competent cells from -80°C (use one tube for each transformation), thaw the cells on ice.
- Add 1-2 μl of DpnI treated PCR product or the template DNA only control into the Dh5acompetent cell tubes, mix by gently tapping the tube several times. DO NOT VORTEX OR PIPETTE UP AND DOWN!!!
- Incubate on ice for 30 minutes.
- Heat shock in a 42°C water bath for 45 seconds.
- Immediately put the tubes back on ice for 2 minutes.
- Add 1 ml of liquid LB media that has been pre-warmed to 37°C into each transformation tube.
- Recover: Shake the tubes in a 37°C shaker at 200 rpm for 60 minutes.
- To pellet the cells, centrifuge the tubes at 5000 rpm for 3 minutes, remove most of the supernatant, leave 150-200 μl, then gently pipette up and down several times to resuspend the cells.
- Transfer cells onto the LB agar plate containing the appropriate antibiotics, spread cells with a glass rod spreader or glass beads.
- Incubate the plate at 37°C overnight,.
- Pick 3-6 colonies, mini-prep plasmids, select the correct clones by sequencing or PCR.
VII. determine the efficiency of the Site-directed mutagenesis using pUC19 control plasmid
To determine the efficiency of the Site-directed mutagenesis method, the pUC19 2.7-kb control plasmid is used to test the efficiency of mutant plasmid formation. The control primers provided in the kit create two point mutations in LacZ-a gene encoded on pUC19. Following DpnI digestion and DH5a transformation, determine the mutagenesis efficiency by counting colony numbers.